PyData London 2016

The Lyst Data Science team was out in force at PyData London 2016 this May. The talks spanned topics in data science, python in science (from lasers to orangutans), plenty of deep learning, and lots of Bayesianism. Here we dive in and pick up on a few of our favourites.
Pete Owlett, “Lessons from 6 months of using Luigi in production”
In this masterful piece of storytelling, Pete took us on a journey from his start as Deliveroo’s first Data Scientist, and the challenges along the way with DIY data engineering.
We were also treated to a brief introduction to the Luigi framework for building up data pipelines.
Thomas Wiecki, “Probabilistic Programming in Data Science with PyMC3”
Probabilistic programming concerns the construction of a probabilistic model in code, with which we may then perform inference. Thomas Wiecki introduced us to the pyMC3 package which exposes a clean API for the specification of such models.
He gave an example of interest from Quantopian: users can submit strategies for algorithmic trading in financial markets, which are backtested against historical data, then paper-traded against the market for a fixed period of time. Given what we have measured about a strategy’s performance against a limited amount of unseen data, how confident can we be that the strategy has an edge over the market? Should it be deployed? pyMC3 enables this kind of modelling to be performed with relative ease.
As a special bonus (and a counter to the Lyst Deep Learning Track), he shared an example of a Bayesian neural network implemented in pyMC3 – see this gist.
Jon Sedar, “Hierarchical Bayesian Modelling with PyMC3 and PySTAN”
Jon Sedar followed up on the above talk with a demonstration of the use of pyMC3 for hierarchical Bayesian models. Such hierarchies arise when we expect relationships in our data: in Jon’s example of car emissions data, we might expect measurements from cars from a given manufacturer to follow a particular distribution, dependent on manufacturer. However, we might hypothesise that these distributions are related, depending on whether manufacturers share a parent owner. This is where hierarchical models come in. In addition to all this, Jon gave a detailed comparison of pyMC3 and pyStan, as well as some pro tips for debugging these kinds of models.
Irwin Zaid, “DyND - Enabling Complex Analytics Across the Language Barrier”
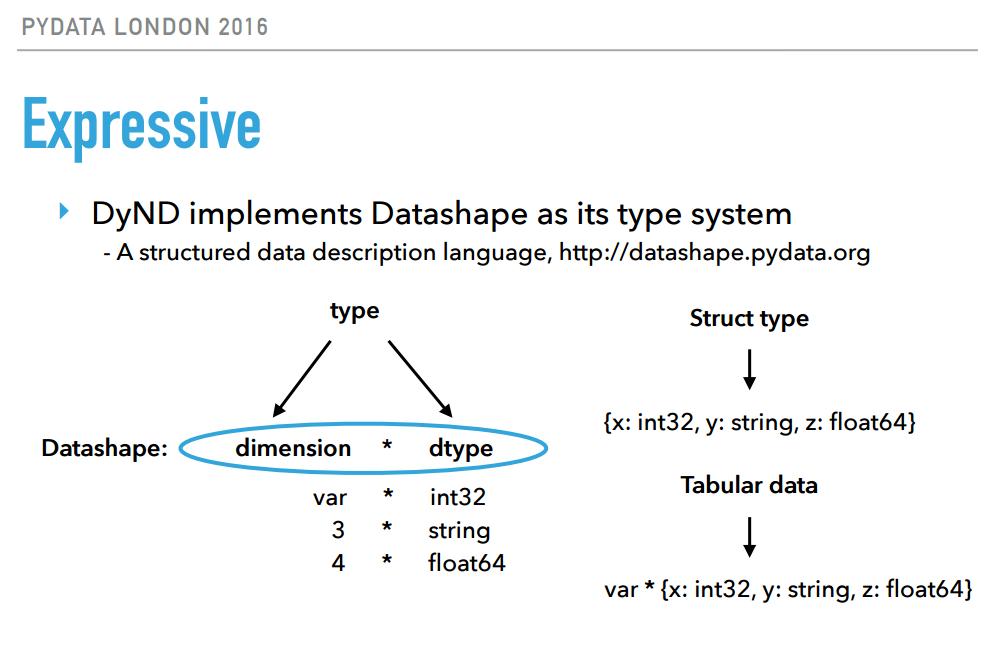
This final pick really appealed to our HPC interests. DyND is a rapidly-developing C++ library for array computation, with python bindings, and plans to support others in the near future. In python, numpy is the present de facto choice for storage and computation on numeric arrays. It was built with HPC requirements in mind: regular arrays of numeric data. Today’s datasets contain missing data, categorical data and ragged arrays, none of which are efficiently handled. DyND and datashape aim to provide a more flexible library for arrays, without a performance penalty. Irwin showed many snippets giving a great sense for how these data types behave. We’re looking forward to seeing where this project is going.
Rich Lewis, “Deep Learning for QSAR”
Rich gave us a dry and humorous introduction to using deep learning for Quantitative Structure Activity Relationship (QSAR). Watch the recording for the full scientific details but essentially QSAR is about mapping molecules to number which characterizes the molecule’s activity in some manner. Rich used a three layer neural network to predict the activity based on a molecular fingerprint and achieved a 50% increase in performance over traditional techniques.
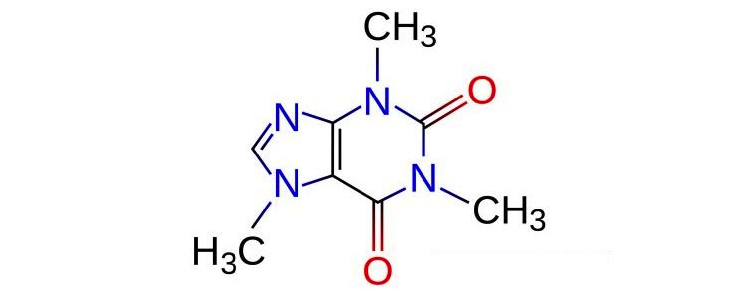
The presentation even included a live demo of caffine binding to A2A adenosine receptor (i.e. Rich taking a sip of coffee).
Frank Kelly and Giles Weaver, “AlzHack: Data Driven Diagnosis of Alzheimer’s Disease”
Frank and Giles gave a great talk on using text data to detect the onset of Alzheimer’s Disease. The talk begins with an introduction into the disease and motivations for finding scalable early detection of the disease. Having previously spoken on using emails from a single sufferer, this follow up talk was based on a new dataset taken from a forum for those affected. By selecting data from sub-forums “I have dementia” and “I have a partner with dementia” they were able to create a dataset with 78,000 posts to analyse.
Frank and Giles carried out a variety of feature extraction techniques and used these with models including Random Forests, SVMs and K-Nearest-Neighbours classifiers. This talk is interesting as an example of applying a variety of text processing techniques for classification, we found the focus of the research to be particularly interesting. Frank finishes the talk asking for anyone interested to get in touch if you have any ideas of other analysis that can be applied.

Dirk Gorissen, “Python vs Orangutan”
It’s always a pleasure to attend a talk by Dirk and this one was no different. Dirk talked about a project he did with Internation Animal Rescue to implant orangutans with tags and track them in the Bornean jungle using a drone. We learnt that orangutans are clever, strong and have no neck. His talk also detailed the much anticipated and obligitory “drone gets stuck in a tree”.
Lysters
In addition, several Lysters spoke at PyData. PyData did a great job of getting the videos up quickly.
Eddie Bell (YouTube, slides) and Calvin Giles (YouTube, slides) gave talks on deep learning in fashion. Sandra Greiss (YouTube, slides) spoke on some applications of machine learning in astronomy. Linda Uruchurtu (YouTube, slides) presented some of her work on survival analysis in python.
If you saw our talks at PyData, or want to get in touch with us, contact us on twitter or leave a comment below! We’re also looking to hire Data Engineers so please do apply if you are looking for a new exciting position.
